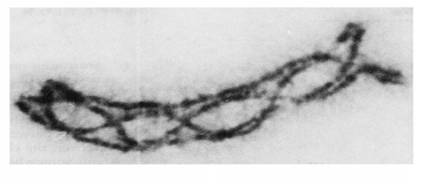
Homologous Recombination (Holliday model and DNA double strand repair model) and Site Specific Recombination
Recombination
- Cleavage and rejoining of DNA molecules to generate new combinations of genes
- There are two types of recombination mechanisms occur in living system
Genetic recombination
Site specific recombination
Genetic Recombination
- The production of gene combinations not found in the parents by the crossing over between homologous chromosomes during meiosis is called genetic recombination.
- Genetic recombination occurs in the germ cells (during meiosis I in oocytes and spermatocytes)
- Also called as homologous recombination as it takes place between homologous chromosomes
- It occurs during crossing over, in which homologous regions of chromosomes are exchanged and genes are shuffled into new combinations.
- It is an extremely important genetic process because it increases genetic variation.
- It is an ubiquitous process occurs in every bacteria and also in eukaryotes
Models of Homologous Recombination
Holliday model (Robin Holliday 1964)
Double strand repair model

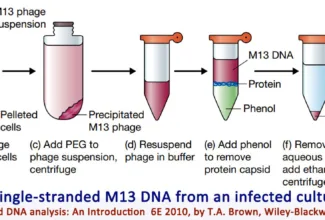
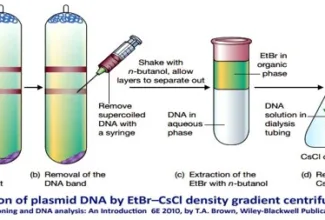

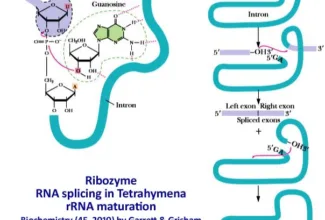
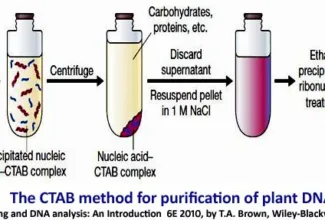
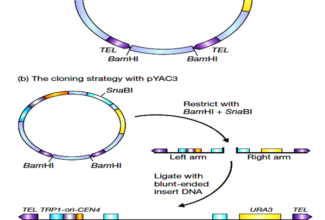


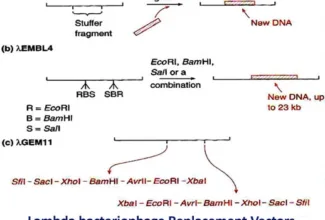







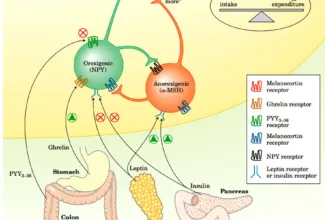

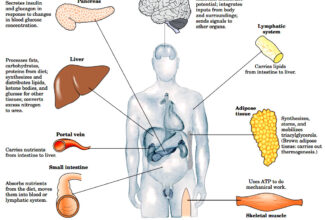
































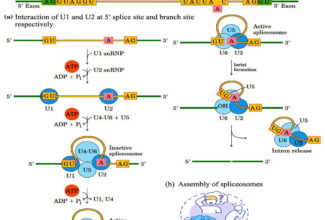























































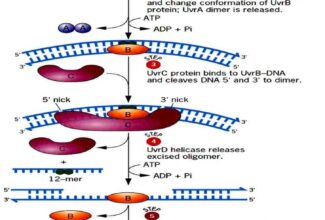





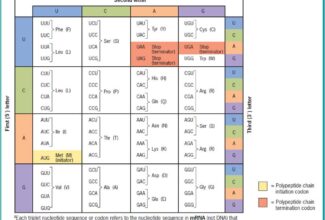







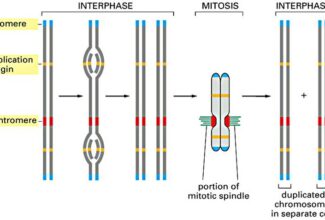











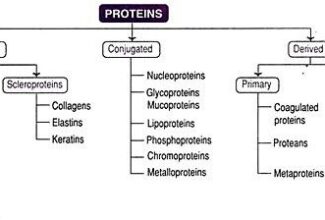


[…] There are 21 MCQs on Homologous Recombination and site specific recombination. the answer key is at the end. To study Homologous Recombination and site specific recombination click on the following linkHomologous Recombination (Holliday model and DNA double strand repair model) and Site Specific Reco… […]